About Our Research
Transcription factors (TFs) regulate gene expression, determining cell fates. We are seeking to predict de novo TF activity in any cell, to perform intentional state transitions (e.g., from diseased to healthy). Our research combines mammalian cell culture models, genetic and epigenetic editing, single cell genomics, large-scale CRISPR screens, and various computational approaches to achieve this goal.
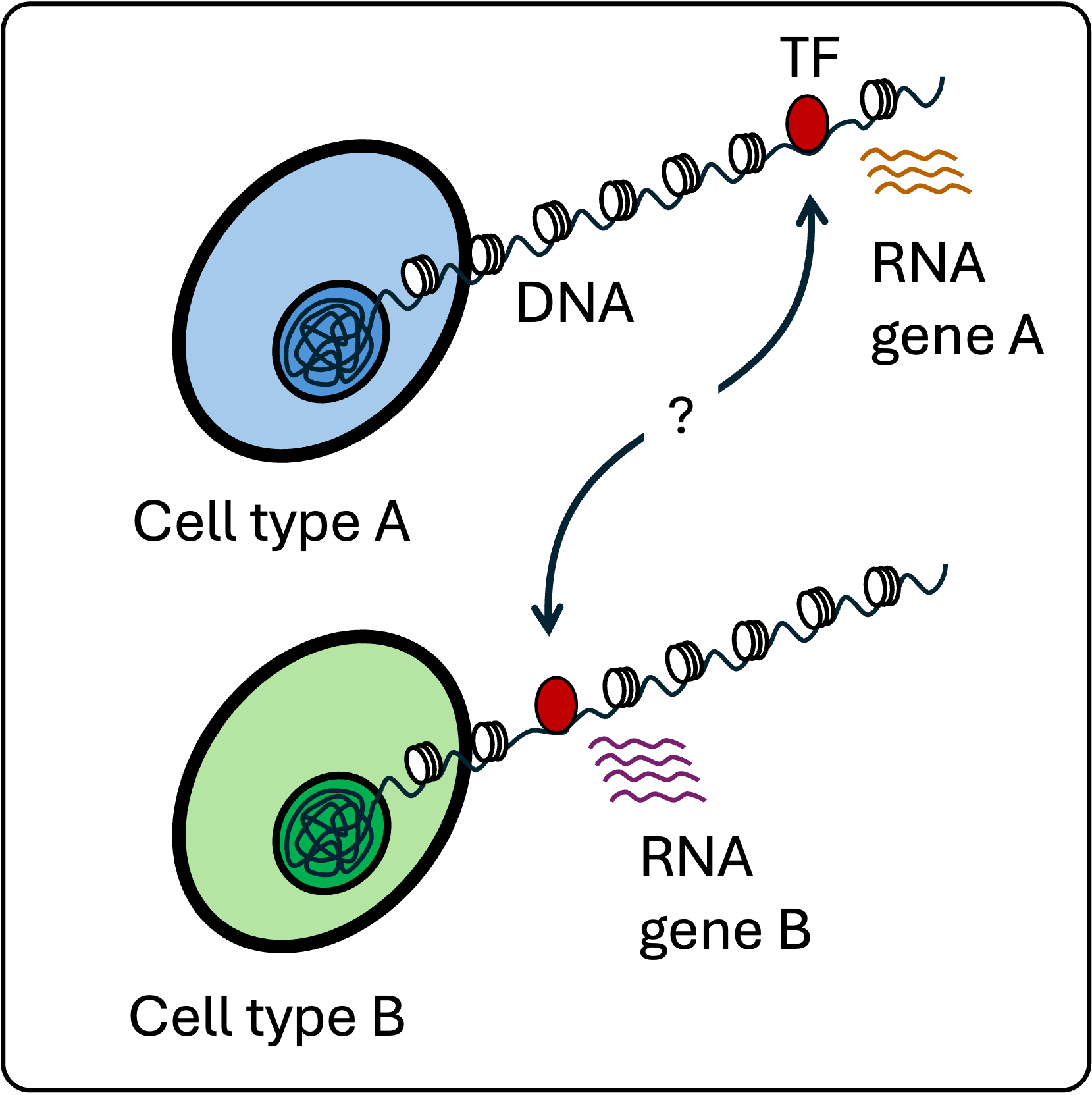
Learning mechanisms of TF binding site selection
TFs bind to specific DNA sequences, but only occupy a subset of potential sites in each cell type. We study the rules governing this cell type-specific binding and its effects.
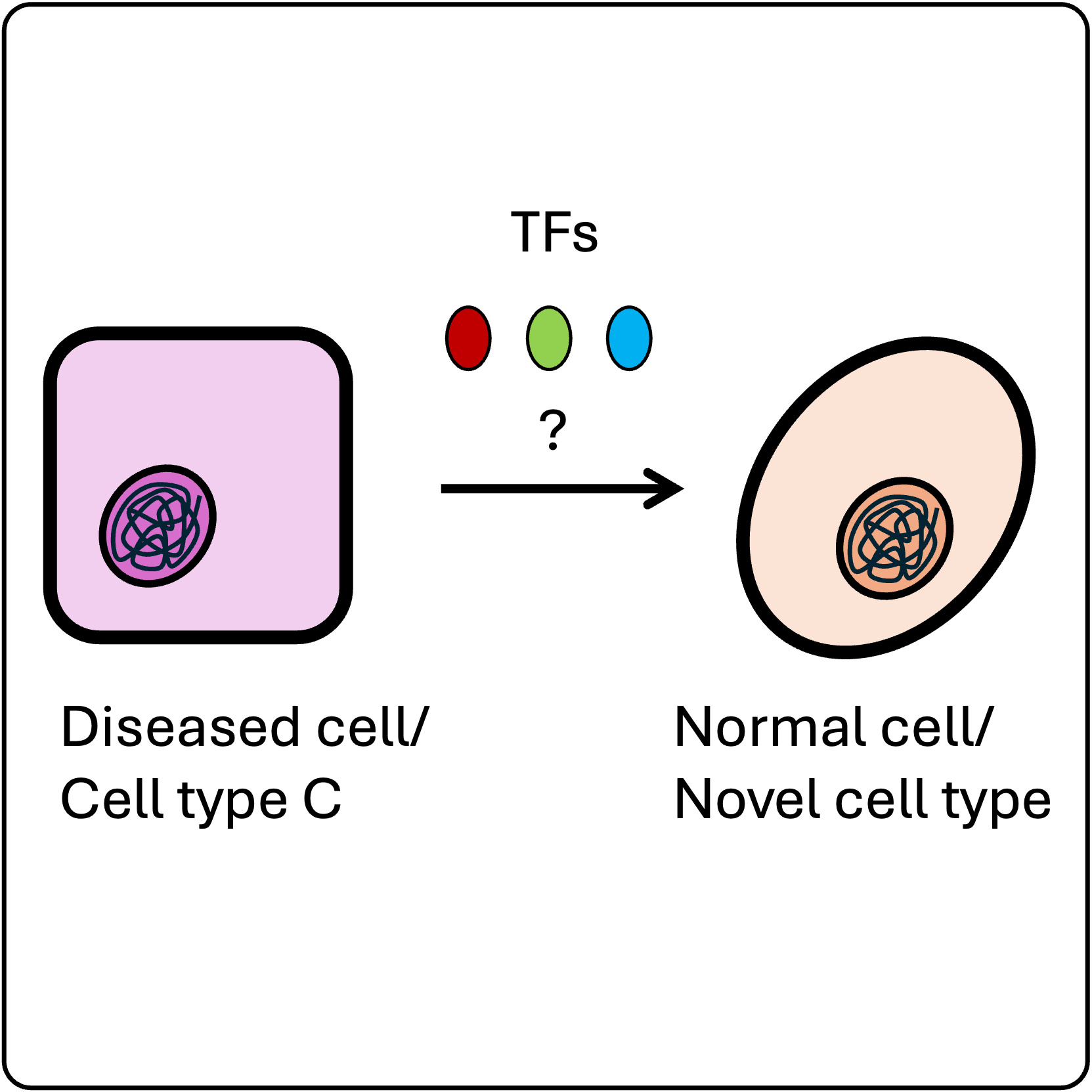
Building predictive models of TF activity
We explore novel stable cell states that can be created with natural TFs. Mapping this opportunity landscape experimentally helps us train deep learning models to predict TF 'recipes' for intentional cell state transitions that do not occur in nature.
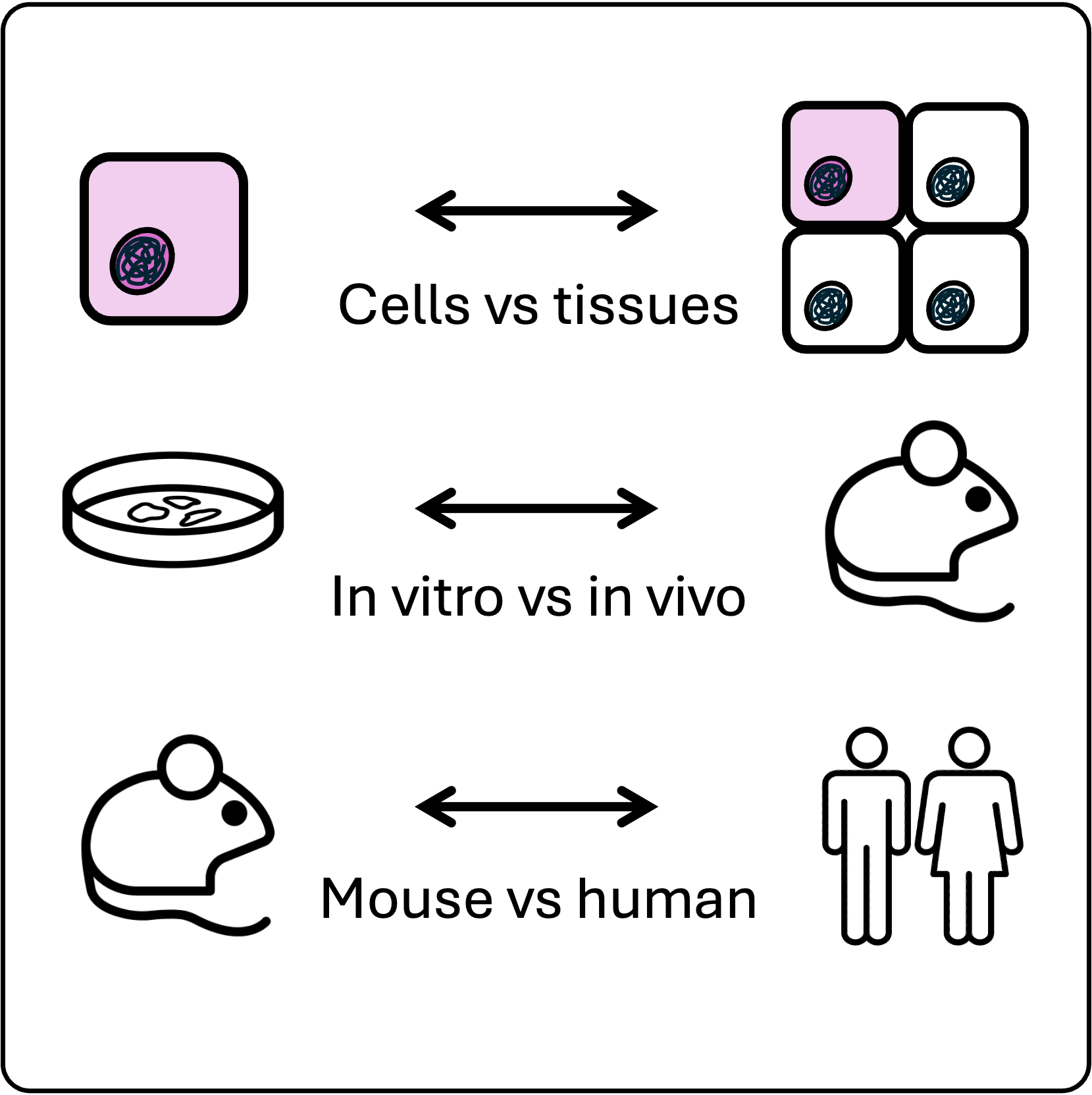
Transferability across scales and organisms
If we were able to predict cell state transitions in patients, this would unlock a new class of therapeutic strategies. To start bridging the gap between the dish and the clinic, we study transferability of our predictions from uni-cellular to multi-cellular systems, across environments and species.